iMutSig - a web application to identify the most similar mutational signature using shiny
Abstract
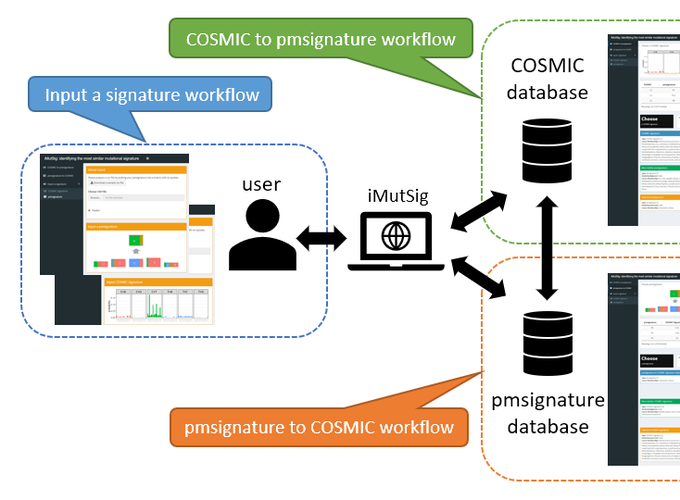
There are two frameworks for characterizing mutational signatures which are commonly used to describe the nucleotide patterns that arise from mutational processes. Estimated mutational signatures from fitting these two methods in human cancer can be found online, in the Catalogue Of Somatic Mutations In Cancer (COSMIC) website or a GitHub repository. The two frameworks make differing assumptions regarding independence of base pairs and for that reason may produce different results. Consequently, there is a need to compare and contrast the results of the two methods, but no such tool currently exists. In this paper, we provide a simple and intuitive interface that allows such comparisons to be easily performed. When using our software, the user may download published mutational signatures of either type. Mutational signatures from the pmsignature data source are expanded to probabilistic vectors of 96-possible mutation types, the same model specification used by COSMIC, and then compared to COSMIC signatures. Cosine similarity measures the extent of signature similarity. iMutSig provides a simple and user-friendly web application allowing researchers to compare signatures from COSMIC to those from pmsignature, and vice versa. Furthermore, iMutSig allows users to input a self-defined mutational signature and examine its similarity to published signatures from both data sources.